LiberTEM UDFs
[1]:
%matplotlib inline
[2]:
import os
import matplotlib.pyplot as plt
import libertem.api as lt
import numpy as np
[3]:
ctx = lt.Context()
Specifying the dataset
Most formats can be loaded using the "auto" type, but some may need additional parameters.
See the loading data section of the LiberTEM docs for details.
[4]:
data_base_path = os.environ.get("TESTDATA_BASE_PATH", "/home/alex/Data/")
[5]:
ds = ctx.load("auto", path=os.path.join(data_base_path, "01_ms1_3p3gK.hdr"))
After loading, some information is available in the diagnostics attribute:
[6]:
ds.diagnostics
[6]:
[{'name': 'Bits per pixel', 'value': '12'},
{'name': 'Data kind', 'value': 'u'},
{'name': 'Layout', 'value': '(1, 1)'},
{'name': 'Partition shape', 'value': '(2075, 256, 256)'},
{'name': 'Number of partitions', 'value': '32'},
{'name': 'Number of frames skipped at the beginning', 'value': 0},
{'name': 'Number of frames ignored at the end', 'value': 0},
{'name': 'Number of blank frames inserted at the beginning', 'value': 0},
{'name': 'Number of blank frames inserted at the end', 'value': 0}]
Standard analyses: virtual detector
A standard analysis to run on 4D STEM data is to apply a virtual detector. Here, we define a ring detector, with radii in pixels:
[7]:
ring = ctx.create_ring_analysis(dataset=ds, ri=60, ro=70)
The analysis can be run with the Context.run method:
[8]:
ring_res = ctx.run(ring, progress=True)
ring_res
[8]:
[<AnalysisResult: intensity>, <AnalysisResult: intensity_log>]
As the analysis mirrors what the web GUI does, we have to access the data using the raw_data attribute, as we would get a viusalized result otherwise. Here we do the visualization ourselves using matplotlib:
[9]:
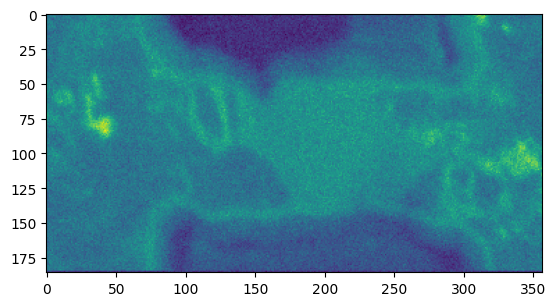
plt.figure()
plt.imshow(ring_res.intensity.raw_data)
[9]:
<matplotlib.image.AxesImage at 0x7fa0146aff90>
Simple UDF definition
User-defined funtions provide a way for you to implement your own data processing functionality. As a very simple example, we define a function that just sums up the pixels of each frame:
[10]:
def sum_of_pixels(frame):
return np.sum(frame)
The easiest way to run this on the data is to use the Context.map function:
[11]:
res_pixelsum_1 = ctx.map(dataset=ds, f=sum_of_pixels, progress=True)
res_pixelsum_1
[11]:
<BufferWrapper kind=nav dtype=float32 extra_shape=()>
The result is of type BufferWrapper, but can be used by any function that expects a numpy array, for example for plotting it:
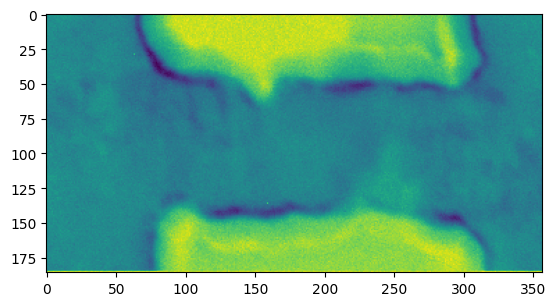
[12]:
plt.figure()
plt.imshow(res_pixelsum_1)
[12]:
<matplotlib.image.AxesImage at 0x7f9ff47d7f90>
The Context.map function is a shortcut for implementing very easy mapping over data, in a frame-by-frame fashion. The longer way of writing this would be as follows:
[13]:
from libertem.udf import UDF
class SumOfPixels(UDF):
def get_result_buffers(self):
return {
'sum_of_pixels': self.buffer(kind='nav', dtype='float32')
}
def process_frame(self, frame):
self.results.sum_of_pixels[:] = np.sum(frame)
This can now be run using the Context.run_udf method:
[14]:
res_pixelsum_2 = ctx.run_udf(dataset=ds, udf=SumOfPixels(), progress=True)
res_pixelsum_2
[14]:
{'sum_of_pixels': <BufferWrapper kind=nav dtype=float32 extra_shape=()>}
The result is now a dict, which maps buffer names, as defined in get_result_buffers, to the BufferWrapper result, so we can use the following to plot the results:
[15]:
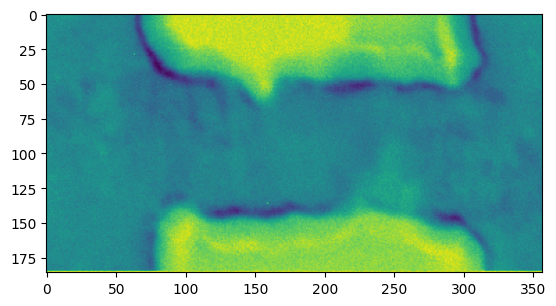
plt.figure()
plt.imshow(res_pixelsum_2['sum_of_pixels'])
[15]:
<matplotlib.image.AxesImage at 0x7f9ff413c090>
extra_shape: more than one result per scan position
[16]:
class StatsUDF(UDF):
def get_result_buffers(self):
return {
'all_stats': self.buffer(kind='nav', dtype='float32', extra_shape=(4,)),
}
def process_frame(self, frame):
self.results.all_stats[:] = (np.mean(frame), np.min(frame), np.max(frame), np.std(frame))
[17]:
res_stats = ctx.run_udf(dataset=ds, udf=StatsUDF(), progress=True)
Result now has an extra dimension, as specified by extra_shape above:
[18]:
res_stats['all_stats'].data.shape
[18]:
(186, 357, 4)
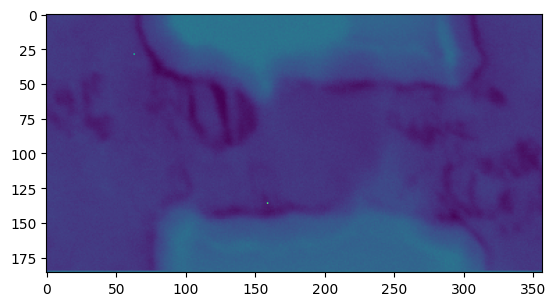
Let’s plot the stddev of each frame:
[19]:
plt.figure()
plt.imshow(res_stats['all_stats'].data[..., 3])
[19]:
<matplotlib.image.AxesImage at 0x7fa054600d90>
kind=”sig” buffers, merge functions
Previously: one result for each scan position
Now: result buffer shaped like the diffraction patterns
We need a merge function to merge the result of one partition into the final result
Different buffer kinds can be combined in a single UDF, so you can combine different operations in a single pass over the data
[20]:
class MaxFrameUDF(UDF):
def get_result_buffers(self):
return {
'maxframe': self.buffer(kind='sig', dtype='float32')
}
def process_frame(self, frame):
# element-wise maximum:
self.results.maxframe[:] = np.maximum(self.results.maxframe, frame)
def merge(self, dest, src):
# src: the maximum observed in the current partition
# dest: the maximum observed in all partitions that were already merged together
dest.maxframe[:] = np.maximum(dest.maxframe, src.maxframe)
[21]:
res_max = ctx.run_udf(dataset=ds, udf=MaxFrameUDF(), progress=True)
[22]:
plt.figure()
plt.imshow(np.log1p(res_max['maxframe']))
[22]:
<matplotlib.image.AxesImage at 0x7fa0342a5950>
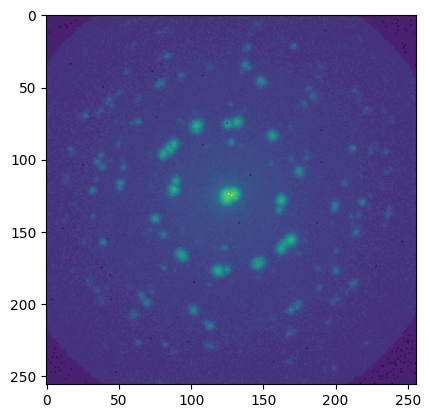
Region of interest
work on a subset of the navigation axes
can be used with all UDFs
useful for working selectively on data, or just reducing the I/O and computational load when implementing a new UDF
defined as a binary mask
Let’s create a mask based on the previously calculated pixel-sum:
[23]:
from skimage.morphology import opening, closing
[24]:
np.min(res_pixelsum_1), np.max(res_pixelsum_1), np.mean(res_pixelsum_1)
[24]:
(np.float32(7762.0), np.float32(12367.0), np.float32(10277.385))
[25]:
mask = res_pixelsum_1.data < np.mean(res_pixelsum_1)
# mask = opening(mask)
mask = closing(opening(mask))
plt.figure()
plt.imshow(mask.astype("float32"))
plt.colorbar()
[25]:
<matplotlib.colorbar.Colorbar at 0x7fa03425f610>
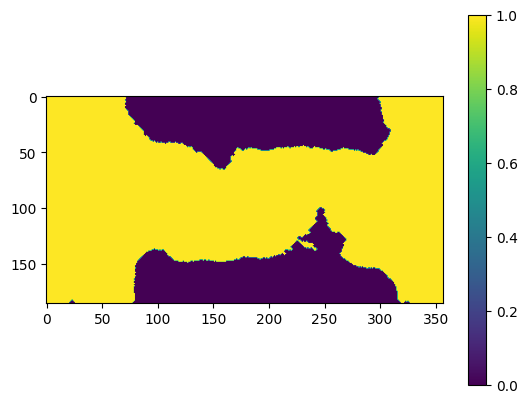
[26]:
res_roi = ctx.run_udf(dataset=ds, udf=StatsUDF(), roi=mask, progress=True)
[27]:
plt.figure()
plt.imshow(np.log1p(res_roi['all_stats'].data[..., 2]))
[27]:
<matplotlib.image.AxesImage at 0x7fa054488210>
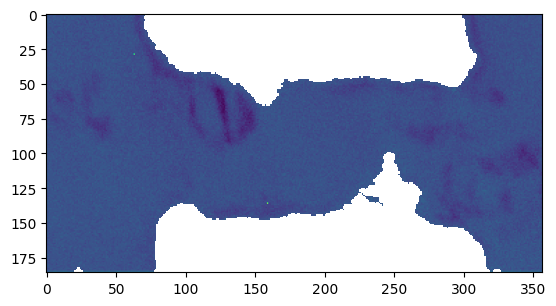
Results with ROI applied
One has to take some care when handling results where a roi was applied; just using the result as a numpy array or accessing the data attribute will give you a result that keeps the whole dataset shape, where the deselected parts are filled with NaN values:
[28]:
res_roi['all_stats'].data.shape
[28]:
(186, 357, 4)
There is a second attribute, raw_data, which will give you a flattenned array of just the results, like numpy would give you for fancy indexing:
[29]:
res_roi['all_stats'].raw_data.shape
[29]:
(45358, 4)
Parameters
Keyword arguments passed to the UDF are made available in self.params on the UDF. A good convention to document your parameters is to put a docstring into your __init__ method and just pass on the values to super().__init__. You can also validate the arguments here.
[30]:
class PixelPicker(UDF):
def __init__(self, coords, *args, **kwargs):
"""
Parameters
----------
coords : Tuple[int]
The coordinates to look at in each frame
"""
if len(coords) != 2:
raise ValueError("invalid coordinates")
super().__init__(*args, coords=coords, **kwargs)
def get_result_buffers(self):
return {
'value_of_pixel': self.buffer(kind='nav', dtype=np.float32)
}
def process_frame(self, frame):
self.results.value_of_pixel[:] = frame[self.params.coords]
[31]:
res_pixel_picker = ctx.run_udf(dataset=ds, udf=PixelPicker(coords=(128, 128)), progress=True)
[32]:
plt.figure()
plt.imshow(res_pixel_picker['value_of_pixel'])
[32]:
<matplotlib.image.AxesImage at 0x7fa054535450>
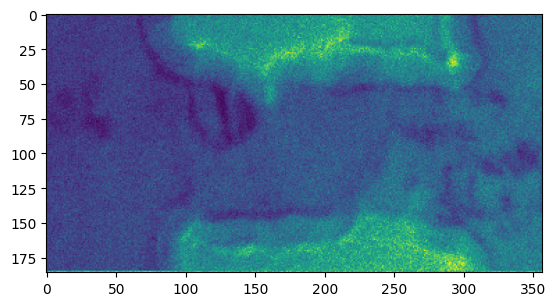
[ ]: