Note
The features described below are experimental and under development.
Dask integration
New in version 0.9.0.
Since version 0.9, LiberTEM supports seamless integration with workflows that
are based on Dask arrays. That means Dask arrays can serve as input for LiberTEM, and
UDF computations can produce Dask arrays. Additionally,
make_dask_array() can create Dask arrays from LiberTEM datasets.
Linking with HyperSpy Lazy Signals
LiberTEM’s Dask integration features can be used to combine features from HyperSpy and LiberTEM in a single analysis workflow:
Scheduler
LiberTEM uses Dask.distributed as the default method to execute tasks.
Unless instructed otherwise, LiberTEM keeps the default Dask scheduler as-is and only
uses the Dask Client internally to make sure existing workflows keep running
as before. However, for a closer integration it can be beneficial to use the same scheduler
for both LiberTEM and other Dask computations. There are several options for that:
Set the LiberTEM Dask cluster as default Dask scheduler:
Use
Context.make_with('dask-make-default')(make_with())Pass
client_kwargs={'set_as_default': True}toconnect()ormake_local()
Use an existing Dask scheduler:
Use
Context.make_with('dask-integration')to start an executor that is compatible with the current Dask scheduler.
Use dask.delayed:
libertem.executor.delayed.DelayedJobExecutorcan return UDF computations as Dask arrays. The scheduler will only be determined whencompute()is called downstream using the normal mechanisms of Dask.
Load datasets as Dask arrays
The make_dask_array() function can generate
a distributed Dask array from a
DataSet using its partitions as blocks. The
typical LiberTEM partition size is close to the optimum size for Dask array
blocks under most circumstances. The dask array is accompanied with a map of
optimal workers. This map should be passed to the compute() method in
order to construct the blocks on the workers that have them in local storage.
from libertem.contrib.daskadapter import make_dask_array
# Construct a Dask array from the dataset
# The second return value contains information
# on workers that hold parts of a dataset in local
# storage to ensure optimal data locality
dask_array, workers = make_dask_array(dataset)
result = dask_array.sum(axis=(-1, -2)).compute(workers=workers)
Load Dask arrays as datasets
LiberTEM datasets can be created from Dask arrays by using
libertem.api.Context.load() with filetype 'dask'. See
Dask for details. The performance can vary and depends on chunking,
executor, I/O method and Dask array creation method. Please contact us or create an Issue for questions, bug reports
and other feedback!
This basic example shows running a UDF on a Dask array:
import dask.array as da
from libertem.udf.sum import SumUDF
# Create a Context that integrates well with
# the current Dask scheduler
ctx = Context.make_with('dask-integration')
a = da.random.random((23, 42, 17, 19))
ds = ctx.load('dask', a, sig_dims=2)
res = ctx.run_udf(dataset=ds, udf=SumUDF())
assert np.allclose(
a.sum(axis=(0, 1)).compute(),
res['intensity'].raw_data
)
See also Executors on how to set up compatible schedulers for both Dask and LiberTEM.
Create Dask arrays with UDFs
Using a DelayedJobExecutor with a
Context lets run_udf
return results as Dask arrays.
In addition to the usual data
and raw_data properties, which
provide results as numpy arrays eagerly, the results are made available
as dask arrays using the attributes
delayed_data
and delayed_raw_data.
The computation is only performed when the
compute() method is called on a Dask array result.
from libertem.api import Context
from libertem.executor.delayed import DelayedJobExecutor
from libertem.udf.sumsigudf import SumSigUDF
ctx = Context(executor=DelayedJobExecutor())
res = ctx.run_udf(dataset=dataset, udf=SumSigUDF())
print(res['intensity'].delayed_data)
dask.array<reshape, shape=(16, 16), dtype=float32, chunksize=(..., ...), chunktype=numpy.ndarray>
This method allows to create large intermediate results of kind='nav'
efficiently in the form of Dask arrays. The partial results can stay as
ephemeral array chunks on the workers only while they are needed for downstream
calculations instead of transporting them through the cluster and instantiating
the full result on the main node.
For calls to run_udf which run multiple UDFs
or have with UDFs which return multiple results, it is strongly recommended
to compute results with a single call to compute(), in order to re-use
computation over the dataset. By default, Dask does not cache intermediate
results from prior runs, so individual calls to compute() require
a complete re-run of the UDFs that were passed to run_udf.
Merge function for Dask array results
Note
See One-step merge (merge_all) for more information.
LiberTEM already uses an efficient default merging method to create Dask arrays
for UDFs that only use kind='nav' buffers and don’t specify their own
merge().
For all UDFs that define their own merge(),
DelayedJobExecutor will use this
existing udf.merge() function to assemble the final results, in the same
way it is used to assemble partial results in each partition. This is carried
out by wrapping the udf.merge() in dask.delayed calls.
However, the user can also specify a
merge_all() method on their UDF that
combines all partial results from the workers to the complete result in a single
step. This allows the DelayedJobExecutor to
produce a streamlined task tree, which then gives Dask greater scope to
parallelise the merge step and reduce data transfers.
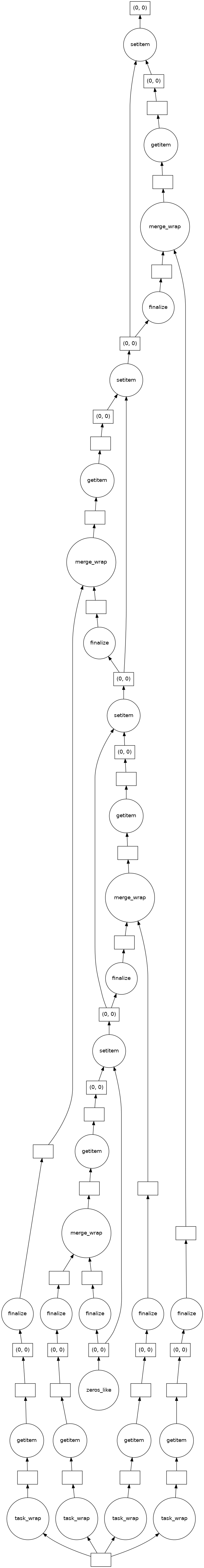
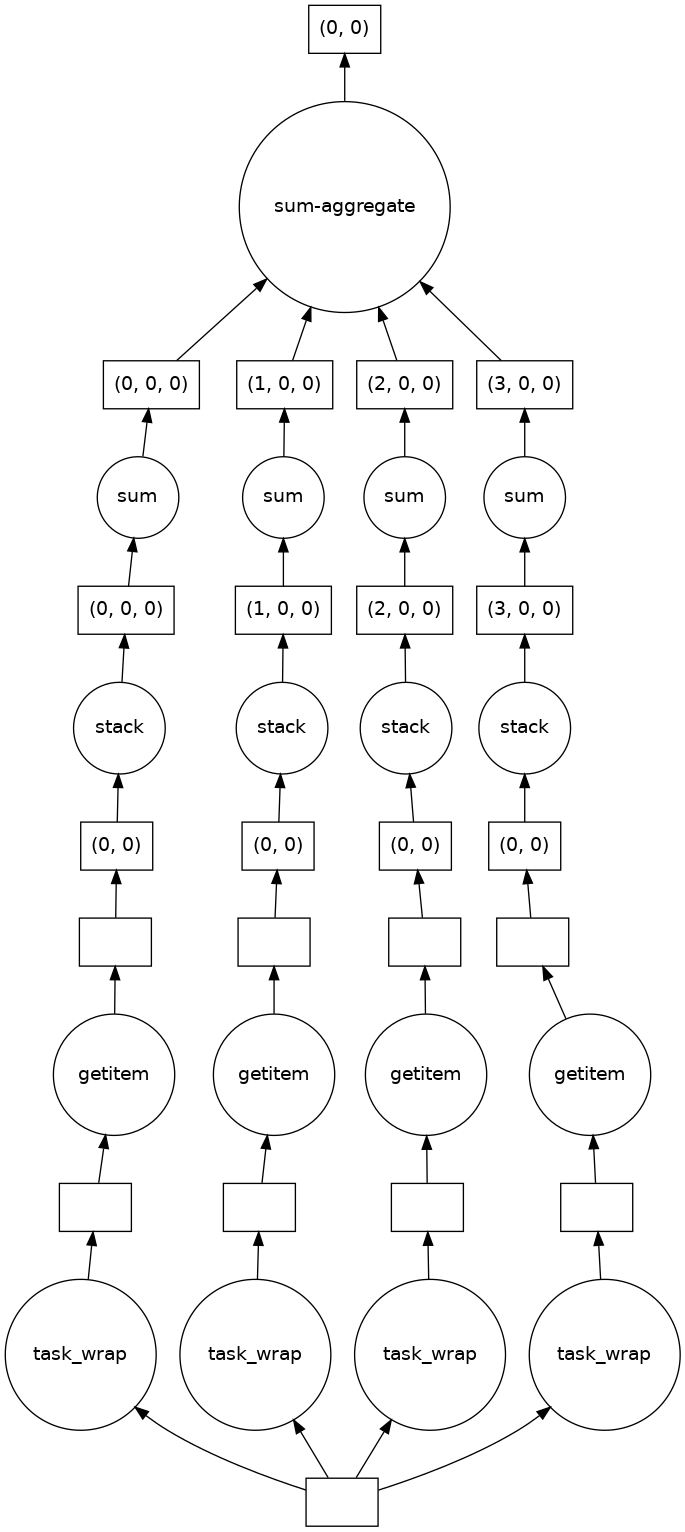
This example shows the task tree first with the built-in wrapper for udf.merge()
and then the stramlined one after defining merge_all(). In tests the streamlined
variant was twice as fast as the built-in wrapper.
class MySumUDF(UDF):
def get_result_buffers(self):
return {
'intensity': self.buffer(kind='sig', dtype=self.meta.input_dtype)
}
def process_tile(self, tile):
self.results.intensity[:] += np.sum(tile, axis=0)
def merge(self, dest, src):
dest.intensity[:] += src.intensity
ctx = Context(executor=DelayedJobExecutor())
result = ctx.run_udf(udf=MySumUDF(), dataset=dataset)
result['intensity'].delayed_data.visualize()
class MySumMergeUDF(MySumUDF):
# Define the merge_all() method for the UDF above
def merge_all(self, ordered_results):
# List and not generator for NumPy dispatch to work
chunks = [b.intensity for b in ordered_results.values()]
# NumPy will dispatch the stacking to the appropriate method
# for the chunks.
# See also https://numpy.org/doc/stable/user/basics.dispatch.html
stack = np.stack(chunks)
# Perform computation on the stacked chunks
# equivalent to the normal merge()
intensity = stack.sum(axis=0)
# Return a dictionary mapping buffer name to new content
return {'intensity': intensity}
result2 = ctx.run_udf(udf=MySumMergeUDF(), dataset=dataset)
result2['intensity'].delayed_data.visualize()
The argument ordered_results is a dictionary of partial results
results for that UDF. The dictionary is keyed by Slice
objects, one for each partition processed, and is ordered in the flat navigation dimension.
Each partial result is itself a dictionary with a key for each result declared in
udf.get_result_buffers(). The ordered_results dictionary is created
such that the merge_all method can safely concatenate the elements in the case
of 'nav'-shaped results. Any applied ROI is automatically taken into account
after the call to merge_all.
The return value from the function must be a dictionary of merged result arrays with the keys matching the declared result buffers. There is, however, no requirement to return merged results for all existing buffers, though any that are missing will not contain results from the computation and are likely to be filled with zeros.
CUDA and scheduling
A native LiberTEM Dask cluster uses resource tags to schedule work on CPUs or
CUDA devices based on an UDF’s capability. For Dask integration a fallback was
implemented that allows running computations that can run on a CPU on a native
LiberTEM Dask cluster without requiring resource tags. However, CUDA-only computations
will require passing the appropriate tags as resources argument to compute().
libertem.executor.delayed.DelayedJobExecutor.get_resources_from_udfs() returns
the appropriate resources for a given set of UDFs based on their capabilities.
Note
At this time the combination of CUDA-requiring UDFs and
DelayedJobExecutor is not well-tested.
At a minimum, the merge process carried out on the main node
will not take place with GPU-backed Dask arrays, though this is
under consideration for the future.