Run UDFs on Merlin live streams
This example shows how to run LiberTEM user-defined functions (UDFs) on Merlin Medipix live data streams. It shows how to customize plotting, and how to integrate LiberTEM-live into your experimental setup.
Make sure to adjust the
NAV_SHAPEbelow to match the scan of the data source!This notebook requires the
bqplotextra of LiberTEM:pip install libertem[bqplot]
Usage with the simulator
If you want to use this with the simulated data source, run a simple Merlin simulator in the background that replays an MIB dataset:
libertem-live-mib-sim ~/Data/default.hdr --cached=MEM --wait-trigger
The --wait-trigger option is important for this notebook to function correctly since that allows to drain the data socket before an acquisition like it is necessary for a real-world Merlin detector.
A suitable MIB dataset can be downloaded at https://zenodo.org/record/5113449.
On Linux, MEMFD is also supported as a cache. Use NONE to deactivate the cache.
[1]:
# Uncomment to use Matplotlib-based plots
# This requires ipympl and allows to capture Matplotlib plots as ipywidgets.
# %matplotlib widget
[2]:
# set this to the host/port where the merlin data server is listening:
MERLIN_DATA_SOCKET = ('127.0.0.1', 6342)
MERLIN_CONTROL_SOCKET = ('127.0.0.1', 6341)
NAV_SHAPE = (128, 128)
[3]:
import logging
import time
import concurrent.futures
import numpy as np
import ipywidgets
from contextlib import contextmanager
[4]:
logging.basicConfig(level=logging.INFO)
[5]:
# Sum all detector frames, result is a map of the detector
from libertem.udf.sum import SumUDF
# Sum up each detector frame, result is a bright field STEM image of the scan area
from libertem.udf.sumsigudf import SumSigUDF
# ImageGL-accelerated plot for fast live display
from libertem.viz.bqp import BQLive2DPlot
# Alternatively a version that uses the slower, but more mature Matplotlib
from libertem.viz.mpl import MPLLive2DPlot
INFO:empyre:Imported EMPyRe V-0.3.1 GIT-e85a58daa6bbd861c3aa1fe26e1d609f376f1adc
[6]:
from libertem_live.api import LiveContext, Hooks
from libertem_live.detectors.merlin import MerlinControl
from libertem_live.udf.monitor import SignalMonitorUDF
[7]:
ctx = LiveContext()
INFO:numba.cuda.cudadrv.driver:init
Camera setup routines
Different from offline processing, the shape, type and content of a dataset is not predetermined in live processing. Instead, the data source has to be configured to supply the desired data. The set_nav() function at the bottom accepts an acquisition object as a parameter to make it easier to configure a matching scan resolution.
[8]:
def merlin_setup(c: MerlinControl, dwell_time=1e-3, depth=6, save_path=None):
print("Setting Merlin acquisition parameters")
# Here go commands to control the camera and the rest of the setup
# to perform an acquisition.
# The Merlin simulator currently accepts all kinds of commands
# and doesn't respond like a real Merlin detector.
c.set('CONTINUOUSRW', 1)
c.set('ACQUISITIONTIME' , dwell_time * 1e3) # Time in miliseconds
c.set('COUNTERDEPTH', depth)
# Soft trigger for testing
# For a real STEM acquisition the trigger setup has to be adapted for the given instrument.
# See the MerlinEM User Manual for more details on trigger setup
c.set('TRIGGERSTART', 5)
c.set('RUNHEADLESS', 1)
c.set('FILEFORMAT', 2) # 0 binary, 2 raw binary
if save_path is not None:
c.set('IMAGESPERFILE', 256)
c.set('FILEENABLE', 1)
c.set('USETIMESTAMPING', 0) # raw format with timestamping is buggy, we need to do it ourselves
c.set('FILEFORMAT', 2) # raw format, less overhead?
c.set('FILEDIRECTORY', save_path)
else:
c.set('FILEENABLE', 0)
print("Finished Merlin setup.")
def microscope_setup(dwell_time=1e-3):
# Here go instructions to set dwell time and
# other scan parameters
# microscope.set_dwell_time(dwell_time)
pass
def set_nav(c: MerlinControl, aq):
height, width = aq.shape.nav
print("Setting resolution...")
c.set('NUMFRAMESTOACQUIRE', height * width)
# Only one trigger for the whole scan with SOFTTRIGGER
# This has to be adapted to the real trigger setup.
# Set to `width` for line trigger and to `1` for pixel trigger.
c.set('NUMFRAMESPERTRIGGER', height * width)
# microscope.configure_scan(shape=aq.shape.nav)
Integration Hooks
A LiberTEM Live acquisition object can include a hooks object, so that LiberTEM Live can set off the acquisition as soon as it has connected to the camera and is ready to receive data. The on_ready_for_data function receives an environment as argument, from which you can access the current acquisition object as the attribute aq.
[10]:
class MerlinHooks(Hooks):
def __init__(self):
self.trigger_result = None
self.pool = concurrent.futures.ThreadPoolExecutor(1)
def on_ready_for_data(self, env):
aq = env.aq
print("Arming Merlin...")
# c is a MerlinControl, will be created in the cell that runs the scan
# below. This arms the detector and sends the acquisition headers.
with c:
c.cmd('STARTACQUISITION')
# microscope.start_scanning()
print("Merlin ready for trigger.")
height, width = aq.shape.nav
# Real-world example: Function call to trigger the scan engine
# that triggers the detector with a hardware trigger to match the scan of the beam.
# This function is blocking until the scan is complete.
# do_scan = lambda: ceos.call.acquireScan(width=width, height=height+1, imageName="test")
# Testing: Use soft trigger
# The emulator can trigger on the 'SOFTTRIGGER' command like the Merlin detector.
def do_scan():
'''
Emulated blocking scan function using the Merlin simulator.
This function doesn't actually block, but it could!
'''
print("Triggering! (do_scan)")
with c:
c.cmd('SOFTTRIGGER')
time.sleep(1) # microscopes can block here
return "stuff" # this result can be queried, once the scan has finished (see last cell)
# The real-world scan function might be blocking. We run it in a thread pool here
# so that `trigger()` returns and the acquisition can start.
fut = self.pool.submit(do_scan)
self.trigger_result = fut
[11]:
conn = ctx.make_connection('merlin').open(
data_host=MERLIN_DATA_SOCKET[0],
data_port=MERLIN_DATA_SOCKET[1],
api_host=MERLIN_CONTROL_SOCKET[0],
api_port=MERLIN_CONTROL_SOCKET[1],
)
[12]:
hooks = MerlinHooks()
aq = ctx.make_acquisition(
conn=conn,
hooks=hooks,
nav_shape=NAV_SHAPE,
frames_per_partition=800,
)
[13]:
udfs = [SumUDF(), SumSigUDF(), SignalMonitorUDF()]
[14]:
LivePlot = BQLive2DPlot
# Uncomment below to use Matplotlib-based plotting
# See also the top of the notebook to select the correct matplotlib backend
# LivePlot = MPLLive2DPlot
p0 = LivePlot(aq, udfs[0])
p1 = LivePlot(aq, udfs[1])
p2 = LivePlot(aq, udfs[2])
[15]:
# NBVAL_IGNORE_OUTPUT
# (output is ignored in nbval run because it somehow doesn't play nice with bqplot)
outputs = []
for p in [p0, p1, p2]:
# Capture the plots to display them in a grid later
output = ipywidgets.Output()
with output:
p.display()
# Some plot-specific tweaks for grid display
if isinstance(p, BQLive2DPlot):
p.figure.fig_margin={'top': 50, 'bottom': 0, 'left': 25, 'right': 25}
p.figure.layout.width = '300px'
p.figure.layout.height = '300px'
elif isinstance(p, MPLLive2DPlot):
p.fig.tight_layout()
p.fig.set_size_inches((3, 3))
p.fig.canvas.toolbar_position = 'bottom'
outputs.append(output)
[16]:
# Show the plot grid
ipywidgets.HBox(outputs)
Sample output
The plots are not preserved when saving the notebook. They look like this:
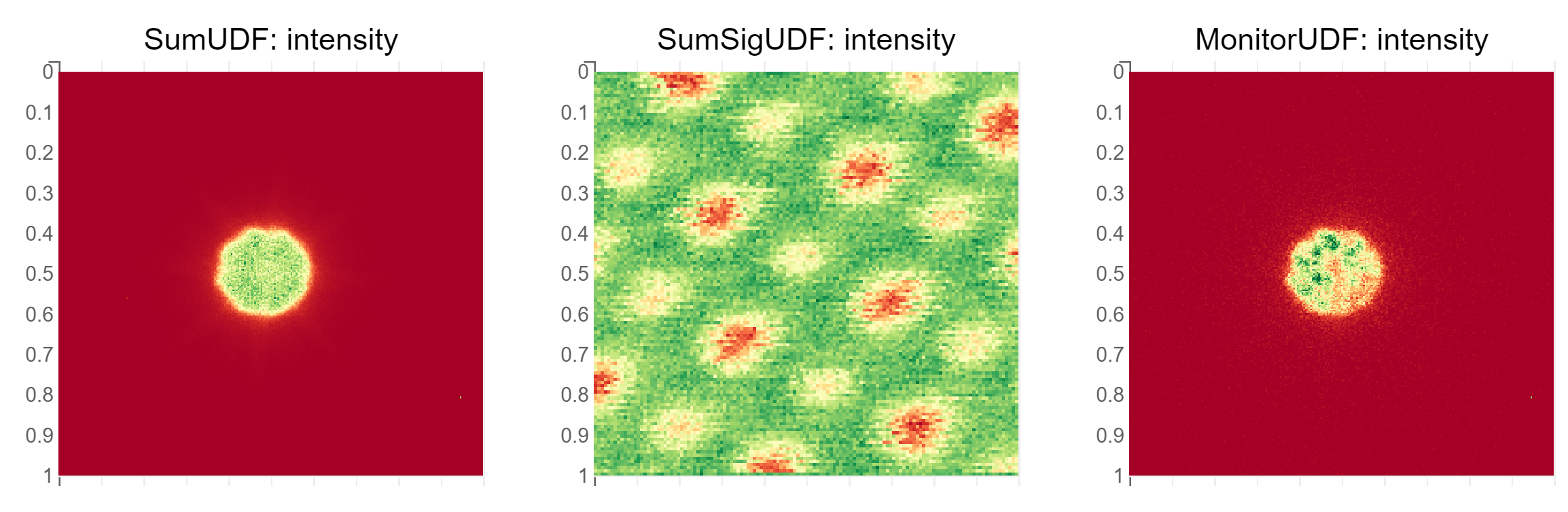
Run one scan
The live plots above are updated with the results
[17]:
c = MerlinControl(*MERLIN_CONTROL_SOCKET)
print("Connecting Merlin control...")
with c:
merlin_setup(c)
microscope_setup()
set_nav(c, aq)
try:
# This will call the trigger function defined above as soon as
# LiberTEM-live is ready to receive data.
ctx.run_udf(dataset=aq, udf=udfs, plots=[p0, p1, p2])
finally:
try:
if hooks.trigger_result is not None:
print("Waiting for blocking scan function...")
print(f"result = {hooks.trigger_result.result()}")
finally:
# Real world:
# microscope.stop_scanning()
pass
print("Finished.")
Connecting Merlin control...
Setting Merlin acquisition parameters
Finished Merlin setup.
Setting resolution...
Arming Merlin...
Merlin ready for trigger.
Triggering! (do_scan)
Waiting for blocking scan function...
result = stuff
Finished.
[ ]: